⬤ Scientists at Tsinghua University have unveiled DrugCLIP, an AI system that could revolutionize early-stage drug discovery. The platform uses deep learning to efficiently pair chemical compounds with human protein targets—one of the biggest bottlenecks in pharmaceutical research.
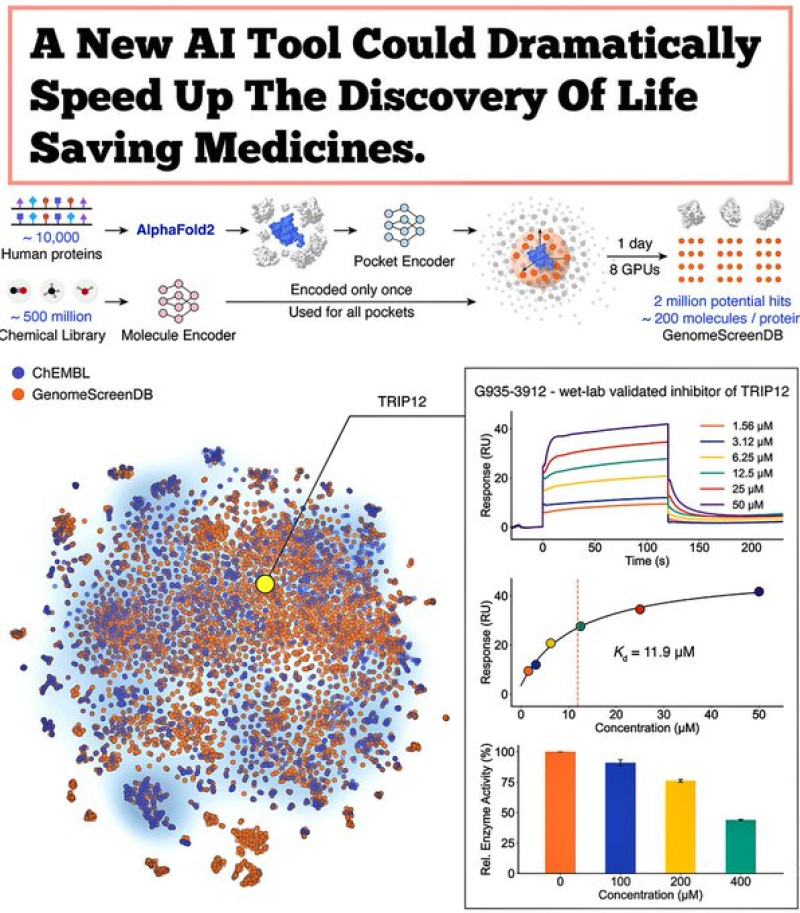
⬤ Here's what makes it powerful: DrugCLIP uses deep contrastive learning to convert both small-molecule structures and protein binding pockets into numerical vectors that can be compared almost instantly. The system screened roughly 500 million molecules against over 10,000 human proteins—about half of all known druggable proteins in the human body. The result? Around 10 trillion molecule-protein evaluations completed in a single day, a throughput that leaves traditional docking simulations in the dust.
⬤ The workflow starts with protein structures generated by AlphaFold2, which are then refined using a custom tool called GenPack to pinpoint binding pockets more accurately. This setup lets DrugCLIP zero in on biologically relevant interaction sites before matching them with candidate molecules. In one breakthrough test, the model identified potential inhibitors for TRIP12—a notoriously difficult-to-target protein linked to cancer and autism. Lab validation confirmed the AI predictions actually worked, showing real inhibitory activity.

⬤ What really matters here is accessibility and scale. All datasets and models are publicly available, meaning research teams worldwide can use this approach without building expensive infrastructure from scratch. By enabling faster, broader exploration of protein-drug interactions, DrugCLIP could cut discovery timelines, lower research costs, and open up new classes of drug targets. As AI embeds itself deeper into biomedical research, systems operating at this scale may fundamentally reshape how new drugs are developed.
 Eseandre Mordi
Eseandre Mordi

 Eseandre Mordi
Eseandre Mordi


